Question: Chapter 21 describes a technique known as
Chapter 21 describes a technique known as Northern blotting that is used to detect RNA transcribed from a particular gene. In this method, a specific RNA is detected using a short segment of cloned DNA as a probe. The DNA probe, which is labeled, is complementary to the RNA that the researcher wishes to detect. After the probe DNA binds to the RNA, the RNA is run on a gel and then visualized as a labeled (dark) band. As shown here, the method of Northern blotting can be used to determine the amount of a particular RNA transcribed in a given cell type. If one type of cell produces twice as much of a particular mRNA as another type of cell does, the band will appear twice as intense. Also, the method can distinguish whether alternative RNA splicing has occurred to produce an RNA that has a different molecular mass.
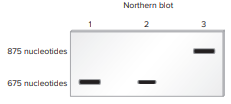
Lane 1 is a sample of RNA isolated from nerve cells.
Lane 2 is a sample of RNA isolated from kidney cells. Nerve cells produce twice as much of this RNA as do kidney cells.
Lane 3 is a sample of RNA isolated from spleen cells. Spleen cells produce an alternatively spliced version of this RNA that is about 200 nucleotides longer than the RNA produced in nerve and kidney cells.
Let’s suppose a researcher is interested in the effects of mutations on the expression of a particular protein-encoding gene in eukaryotes. The gene has one intron that is 450 nucleotides long. After this intron is removed from the pre-mRNA, the mRNA transcript is 1100 nucleotides in length. Diploid somatic cells have two copies of this gene. Make a drawing that shows the expected results of a Northern blot using mRNA from the cytosol of somatic cells, which were obtained from the following individuals:
Lane 1: A normal individual
Lane 2: A homozygote for a deletion that removes the −50 to −100 region of the gene that encodes this mRNA
Lane 3: A heterozygote in which one gene is normal and the other gene has a deletion that removes the −50 to −100 region
Lane 4: A homozygote for a mutation that introduces an early stop codon into the middle of the coding sequence of the gene
Lane 5: A homozygote for a two-nucleotide deletion that removes the AG sequence at the 3′ splice site
Transcribed Image Text:
Northern blot 1 2 3 875 nucleotides 675 nucleotides
> If these two genes were very far apart on the bacterial chromosome, how would the results have been different?
> Describe how genetic transfer can explain the growth of colonies on the middle plate.
> Does mitotic recombination occur in a gamete (sperm or egg cell) or in a somatic cell?
> How do the wobble rules affect the total number of different tRNAs that are needed to carry out translation?
> What phenomenon explains why the maximum percentage of recombinant offspring does not exceed 50%?
> Which types of offspring are found in excess in the F2 generation, based on Mendel’s law of independent assortment?
> DNA replication is fast, virtually error-free, and coordinated with cell division. Discuss which of these three features you think is the most important.
> Explain why all of the offspring in the F2 generation are dextral even though some of them are dd.
> Discuss the types of RNA transcripts and the functional roles they play. Why do you think some RNAs form complexes with protein subunits?
> Based on your knowledge of introns and pre-mRNA splicing, discuss whether or not you think alternative splicing fully explains the existence of introns. Can you think of other possible reasons to explain their existence?
> Compare and contrast DNA replication in bacteria and eukaryotes.
> The complementarity of its two strands is the underlying reason that DNA can be faithfully copied. Propose alternative chemical structures that could be faithfully copied.
> Discuss and make a list of the similarities and differences between bacterial and eukaryotic chromosomes.
> The prevalence of highly repetitive sequences seems rather strange to many geneticists. Do they seem strange to you? Why or why not? Discuss whether or not you think they have an important function.
> Bacterial and eukaryotic chromosomes are very compact. Discuss the advantages and disadvantages of a compact chromosomal structure.
> Cell biology textbooks often discuss cellular proteins encoded by genes that are members of a gene family. Examples of such proteins include myosins and glucose transporters. Look through a cell biology textbook and identify some proteins encoded by memb
> How might you provide evidence that DNA is the genetic material in mice?
> Try to propose structures for a genetic material that are substantially different from the double helix. Remember that the genetic material must have a way to store information and a way to be faithfully replicated.
> Which allele is an example of a loss-of-function allele?
> What are the two key functional sites of a tRNA molecule?
> Besides the ones mentioned in this textbook, look for other examples of variations in euploidy. Perhaps you might look in more advanced textbooks concerning population genetics, ecology, etc. Discuss the phenotypic consequences of these changes.
> A chromosome that was involved in a reciprocal translocation also has an inversion. In addition, the cell contains two normal chromosomes. Make a drawing that shows how these chromosomes will pair during metaphase of meiosis I. A B K M M DE D Invert
> Consider this cross in pea plants: Tt Rr yy Aa × Tt rr Yy Aa, where T = tall, t = dwarf, R = round, r = wrinkled, Y = yellow, y = green, A = axial, a = terminal. What is the expected phenotypic outcome of this cross? Have one group of students solve this
> Mendel studied seven traits in pea plants, and the garden pea happens to have seven different chromosomes. It has been pointed out that Mendel was very lucky not to have conducted crosses involving two traits governed by genes that are closely linked on
> Of the three types of genetic transfer, discuss which one(s) is/are more likely to occur between members of different species. Discuss some of the potential consequences of interspecies genetic transfer.
> Discuss the advantages of the genetic analysis of bacteria. Make a list of the types of allelic differences among bacteria that are suitable for genetic analyses.
> In Chapter 3, we discussed the idea that the X and Y chromosomes have a few genes in common. These genes are inherited in a pseudoautosomal pattern. With this phenomenon in mind, discuss whether or not the X and Y chromosomes are really distinct linkage
> In mice, a dominant allele that causes a short tail is located on chromosome 2. On chromosome 3, a recessive allele causing droopy ears is 6 mu away from another recessive allele that causes a flaky tail. A recessive allele that causes a jerker (uncoordi
> Discuss the principles of the chromosome theory of inheritance. Which principles were deduced via light microscopy, and which were deduced from crosses? What modern techniques could be used to support the chromosome theory of inheritance?
> According to the endosymbiosis theory, mitochondria and chloroplasts are derived from bacteria that took up residence within eukaryotic cells. At one time, prior to being taken up by eukaryotic cells, these bacteria were free-living organisms. However, w
> Which of these three scenarios explains overdominance with regard to the sickle cell allele? From Figure 4.8: Pathogen can successfully Pathogen cannot successfully propagate. propagate. A1A1 A1A2 Normal homozygote (sensitive to infection) Heterozyg
> Recessive maternal effect genes are identified in flies (for example) when a phenotypically normal mother cannot produce any normal offspring. Because all of the offspring are dead, this female fly cannot be used to produce a strain of heterozygous flies
> In oats, the color of the chaff is determined by a two-gene interaction. When a true-breeding black chaff plant was crossed to a true-breeding white chaff plant, the F1 generation was composed of all black chaff plants. When the F1 offspring were crossed
> Let’s suppose a gene exists as a functional wild-type allele and a nonfunctional mutant allele. At the organism level (i.e., at the level of visible traits), the wild-type allele is dominant. In a heterozygote, discuss whether dominance occurs at the cel
> A diploid eukaryotic cell has 10 chromosomes (5 per set). As a group, take turns having one student draw the cell as it would look during a phase of mitosis, meiosis I, or meiosis II; then have the other students guess which phase it is.
> In Figure 3.18, Morgan obtained a white-eyed male fly in a population containing many red-eyed flies that he thought were true-breeding. As mentioned in the experiment, he crossed this fly with several red-eyed females, and all the offspring had red eyes
> Consider this four-factor cross: Tt Rr yy Aa × Tt RR Yy aa, where T = tall, t = dwarf, R = round, r = wrinkled, Y = yellow, y = green, A = axial, a = terminal. What is the probability that the first three plants will have round seeds? What is the easiest
> A cross was made between two pea plants, TtAa and Ttaa, where T = tall, t = dwarf, A = axial, and a = terminal. What is the probability that the first three offspring will be tall with axial flowers or dwarf with terminal flowers and the fourth offspring
> Discuss how variation in chromosome number has been useful in agriculture.
> Which events during translation involve molecular recognition between base sequences within different RNAs? Which events involve recognition between different protein molecules?
> Discuss and make a list of the similarities and differences in the events that occur during the initiation, elongation, and termination stages of transcription (see Chapter 12) and translation discussed in this chapter.
> Why does the heterozygote have an advantage?
> Discuss why you think the ribosomes need to contain so many proteins and rRNA molecules. Does it seem like a waste of cellular energy to make such a large structure so that translation can occur?
> Consider how histone proteins bind to DNA and then explain why a high salt concentration can remove histones from DNA (as shown in Figure 10.18b). From Figure 10.18b: 2 jum DNA strand Scaffold.
> If you were given a sample of chromosomal DNA and asked to determine if it is bacterial or eukaryotic, what experiment would you perform, and what would be your expected results?
> Researchers are often interested in focusing their attention on the transcription of protein-encoding genes in eukaryotes. Such researchers want to study mRNA. One method that is used to isolate mRNA is column chromatography. (Note: See Appendix A for a
> The technique of DNase I footprinting is described in Chapter 21. If a protein binds over a region of DNA, it will protect the DNA in that region from digestion by DNase I. To carry out a DNase I footprinting experiment, a researcher has a sample of a cl
> As described in Chapter 21 and in experimental question E3, an electrophoretic mobility shift assay can be used to determine if a protein binds to DNA. This method can also determine if a protein binds to RNA. For each of the following combinations, woul
> An electrophoretic mobility shift assay (EMSA) can be used to study the binding of proteins to a segment of DNA. This method is described in Chapter 21. When a protein binds to a segment of DNA, it slows the movement of the DNA through a gel, so the DNA
> A research group has sequenced the cDNA and genomic DNA for a particular gene. The cDNA is derived from mRNA, so it does not contain introns. Here are the DNA sequences. cDNA: 5′–ATTGCATCCAGCGTATACTATCTCGGGCCCAATTAATG
> Another technique described in Chapter 21 is polymerase chain reaction (PCR) which is based on our understanding of DNA replication. In this method, a small amount of double-stranded template DNA is mixed with a high concentration of primers. Nucleotides
> At which level is incomplete dominance more likely to be observed—at the molecular/cellular level or at the organism level?
> The technique of dideoxy sequencing of DNA is described in Chapter 21. The technique relies on the use of dideoxyribonucleotides. A dideoxyribonucleotide has a hydrogen atom attached to the 3′ carbon atom instead of a hydroxyl (−− OH) group. When a dideo
> As described in Table 11.3, what is the difference between a rapidstop and a slow-stop mutant? What are different roles of the proteins that are defective in rapid-stop and slow-stop mutants? From Figure 11.3: Examples of ts Mutants Involved in DNA
> Figure 11.4b shows an autoradiograph of a replicating bacterial chromosome. If you analyzed many replicating chromosomes, what types of information could you learn about the mechanism of DNA replication? From Figure 11.4b: FReplication fork Replicat
> An absentminded researcher follows the steps of Figure 11.3, and when the gradient is viewed under UV light, the researcher does not see any bands at all. Which of the following mistakes could account for this observation? Explain how. The researcher fo
> Answer the following questions pertaining to the experiment of Figure 11.3. A. What would be the expected results if the Meselson and Stahl experiment were carried out for four or five generations? B. What would be the expected results of the Meselson
> Let’s suppose you have isolated chromatin from some bizarre eukaryote with a linker region that is usually 300–350 bp in length. The nucleosome structure is the same as in other eukaryotes. If you digested this eukaryotic organism’s chromatin with a high
> When chromatin is treated with a salt solution of moderate concentration, the linker histone H1 is removed. A higher salt concentration removes the rest of the histone proteins. If the experiment of Figure 10.11 was carried out after the DNA was treated
> In Noll’s experiment of Figure 10.11, explain where DNase I cuts the DNA. Why were the bands on the gel in multiples of 200 bp at lower DNase I concentrations? From Figure 10.11: Experimental level Conceptual level 1. Incubate the
> We seem to know more about the structure of eukaryotic chromosomal DNA than bacterial DNA. Discuss why you think this is so, and list several experimental procedures that have yielded important information concerning the compaction of eukaryotic chromati
> Let’s suppose you have isolated DNA from a cell and viewed it under a microscope. It looks supercoiled. What experiment would you perform to determine if it is positively or negatively supercoiled? In your answer, describe your expected results. You may
> At the molecular level, what is the explanation for why the four-o’clock flowers are pink instead of red?
> Two circular DNA molecules, which we can call molecule A and molecule B, are topoisomers of each other. When viewed under the electron microscope, molecule A appears more compact than molecule B. The level of gene transcription is much lower for molecule
> Acridine orange is a chemical that inhibits the replication of F-factor DNA but does not affect the replication of chromosomal DNA, even if the chromosomal DNA contains an Hfr. Let’s suppose that you have an E. coli strain that is unable to metabolize la
> A female fruit fly has one normal X chromosome and one X chromosome with a deletion. The deletion occurred in the middle of the X chromosome and removed about 10% of the entire length of the X chromosome. Suppose you stained and observed the chromosomes
> What are G bands? Discuss how G bands are useful in the analysis of chromosome structure.
> Gierer and Schramm exposed plant tissue to purified RNA from tobacco mosaic virus, and the plants developed the same types of lesions as if they had been exposed to the virus itself. What would be the results if the RNA was treated with DNase, RNase, or
> With regard to Chargaff’s experiment described in Figure 9.10, answer the following: A. What is the purpose of paper chromatography? B. Explain why it is necessary to remove the bases in order to determine the base composition of DNA.
> The type of model building used by Pauling and by Watson and Crick involved the use of ball-and-stick units. Now we can do model building on a computer screen. Even though you may not be familiar with this approach, discuss potential advantages of using
> An interesting trait that some bacteria exhibit is resistance to being killed by antibiotics. For example, certain strains of bacteria are resistant to tetracycline, whereas other strains are sensitive to tetracycline. Describe an experiment you would ca
> With regard to the experiment described in Figure 9.2, answer the following: A. List several possible reasons why only a small percentage of the type R bacteria was converted to type S. B. Explain why an antibody was used to remove the bacteria that we
> Genetic material acts as a blueprint for an organism’s traits. Explain how Griffith’s experiments indicated that genetic material was being transferred to the type R bacteria.
> What are the two main factors that determine an organism’s traits?
> It is an exciting time to be a plant breeder because so many options are available for the development of new types of agriculturally useful plants. Let’s suppose you wish to develop a seedless tomato that can grow in a very hot climate and is resistant
> Describe the steps you would take to produce a tetraploid plant from a diploid plant.
> Describe how colchicine can be used to alter chromosome number.
> With regard to the analysis of chromosome structure, explain the experimental advantage that polytene chromosomes offer. Discuss why changes in chromosome structure are more easily detected in polytene chromosomes than in ordinary chromosomes.
> Let’s suppose a researcher conducted comparative genomic hybridization (see Figure 8.9) and accidentally added twice as much DNA from normal cells (labeled with red fluorescence) relative to DNA from cancer cells. What ratio of green-to-red fluorescence
> What is the main goal of comparative genome hybridization? Explain how the ratio of green to red fluorescence provides information about chromosome structure.
> An Hfr strain that is hisE+ and pheA+ was mixed with a strain that is hisE− and pheA−. The conjugation was interrupted and the percentage of recombinants for each gene was determined by streaking on a medium that lacke
> As mentioned in question 2 of More Genetic TIPS, origins of transfer can be located in many different locations, and their direction of transfer can be clockwise or counterclockwise. Let’s suppose a researcher conjugated six different H
> In your laboratory, you have an F− strain of E. coli that is resistant to streptomycin and is unable to metabolize lactose, but it can metabolize glucose. Therefore, this strain can grow on a medium that contains glucose and streptomycin, but it cannot g
> In a conjugation experiment, what is meant by the time of entry? How is the time of entry determined experimentally?
> Which individual(s) in this pedigree exhibit(s) the effect of incomplete penetrance?
> What is an interrupted mating experiment? What type of experimental information can be obtained from this type of study? Why is it necessary to interrupt mating?
> Explain how a U-tube apparatus can distinguish between genetic transfer involving conjugation and genetic transfer involving transduction. Do you think a U-tube could be used to distinguish between transduction and transformation?
> In the experiment of Figure 7.1, Lederberg and Tatum could not discern whether met+ bio+ genetic material was transferred to the met− bio− thr+ leu+ thi+ strain or if thr+ leu+ thi+ genetic material was transferred to
> In the experiment of Figure 7.1, a met− bio− thr+ leu+ thi+ cell could become met+ bio+ thr+ leu+ thi+ by a (rare) double mutation that converts the met− bio− genes into met+ bio+. L
> Two genes, designated A and B, are located 10 mu from each other. A third gene, designated C, is located 15 mu from B and 5 mu from A. The parental generation consisting of AA bb CC and aa BB cc individuals were crossed to each other. The F1 heterozygote
> Two genes are located on the same chromosome and are known to be 12 mu apart. An AABB individual was crossed to an aabb individual to produce AaBb offspring. The AaBb offspring were then testcrossed to aabb individuals. A. If the testcross produces 1000
> In Morgan’s three-factor crosses of Figure 6.3, he realized that crossing over was more frequent between the eye color and wing length genes than between the body color and eye color genes. Explain how he determined this. From Figure 6
> If two genes are more than 50 mu apart, how would you ever be able to show experimentally that they are located on the same chromosome?
> Explain why the percentage of recombinant offspring in a testcross is a more accurate measure of map distance when two genes are close together. When two genes are far apart, is the percentage of recombinant offspring an underestimate or overestimate of
> In your own words, explain why a testcross cannot produce more than 50% recombinant offspring. When a testcross does produce 50% recombinant offspring, what does this result mean?
> Does a PP individual produce more of the protein encoded by the P gene than is necessary for the purple color?
> Explain the rationale behind a testcross. Is it necessary for one of the parents to be homozygous recessive for the genes of interest? In the heterozygous parent of a testcross, must all of the dominant alleles be linked on the same chromosome and all of
> In the experiment of Figure 6.6, Stern followed the inheritance pattern in which females carried two abnormal X chromosomes to correlate genetic recombination with the physical exchange of chromosome pieces. Is it necessary to use a strain carrying two a
> Three recessive traits in garden pea plants are as follows: yellow pods are recessive to green pods, bluish green seedlings are recessive to green seedlings, creeper (a plant that cannot stand up) is recessive to normal. A true-breeding normal plant with
> A sex-influenced trait is dominant in males and causes bushy tails. The same trait is recessive in females. Fur color is not sex influenced. Yellow fur is dominant to white fur. A true-breeding female with a bushy tail and yellow fur was crossed to a whi

