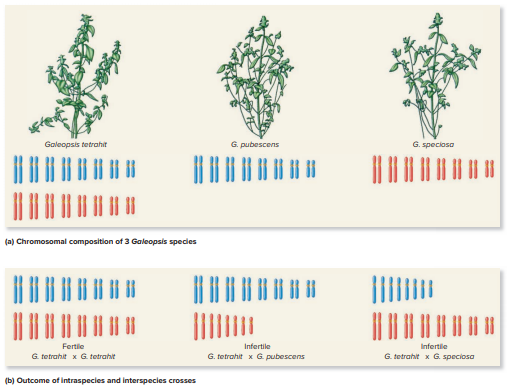
Question: In Chapter 23, a technique called fluorescence
In Chapter 23, a technique called fluorescence in situ hybridization (FISH) is described. In this method, a labeled piece of DNA is hybridized to a set of chromosomes. Let’s suppose that you cloned a piece of DNA from G. pubescens (see Figure 29.4) and used it as a labeled probe for in situ hybridization. What would you expect to happen if this DNA probe were hybridized to the G. speciosa or G. tetrahit chromosomes? Describe the expected results.
From Figure 29.4:
Transcribed Image Text:
Goleopsis tetrahit G. pubescens G. speciosa (a) Chromosomal composition of 3 Galeopsis species I Infertile G. tetrahit x G. speciosa Fertile Infertile G. tetrahit x G. tetrahit G. tetrahit x G pubescens (b) Outcome of intraspecies and interspecies crosses
> Discuss the times in a person’s life when it is most important to avoid mutagens. Which parts of a person’s body should be the most highly protected from mutagens?
> In E. coli, a variety of mutator strains have been identified in which the spontaneous rate of mutation is much higher than in normal strains. Make a list of the types of abnormalities that could cause a strain of bacteria to become a mutator strain. Whi
> Browse the Internet to determine the drugs that are used to treat people with AIDS. Which proteins do these drugs affect? Discuss how an understanding of the HIV reproductive cycle has been helpful in developing treatments for AIDS.
> Certain environmental conditions such as exposure to UV light are known to activate lysogenic λ prophages and cause them to progress into the lytic cycle. UV light initially causes the repressor protein to be proteolytically degraded. Make a flow diagram
> Discuss the properties of emerging viruses. What are the challenges associated with combating them?
> Discuss the advantages and disadvantages of genetic regulation at the different points identified in Figure 14.1. From Figure 14.1: REGULATION OF GENE EXPRESSION Gene Transcription Genetic regulatory proteins bind to the DNA and control the rate of
> Go to the PubMed website and do a search using the words noncoding RNA and disease. Scan through the journal articles you retrieve and make a list of the roles that ncRNAs may play in human diseases.
> Review the concept of an RNA world described in Section 17.1. Discuss which ncRNAs described in Table 17.1 may have arisen during the RNA world, and which probably arose after the modern DNA/RNA/protein world came into being. From Table 17.1: Exampl
> Discuss the similarities and differences of phenotypic variations that are caused by epigenetic gene regulation versus variation in gene sequences (epigenetics versus genetics).
> Go to the PubMed website and search using the words epigenetic and cancer. Scan through the journal articles you retrieve, and make a list of environmental agents that may cause epigenetic changes that contribute to cancer.
> Which form of HIV RNA, fully spliced, incompletely spiced, or unspliced, is needed during the early stage of the synthesis of HIV components?
> Enhancers can occur almost anywhere in DNA and affect the transcription of a gene. Let’s suppose you have a gene cloned on a piece of DNA, and the DNA fragment is 50,000 bp in length. Using cloning methods described in Chapter 21, you can cut out short s
> Explain how DNA methylation could be used to regulate gene expression in a tissue-specific way. When and where would de novo methylation occur, and when would demethylation occur? What would occur in the cells that give rise to eggs and sperm?
> Looking at Figure 14.10, discuss possible “molecular ways” that the cAMP-CAP complex and lac repressor may influence RNA polymerase function. In other words, try to explain how the bending and looping in DNA may affect
> Do you think that Darwin would object to the neutral theory of evolution?
> Compare the forms of speciation that are slow to those that occur more rapidly. Make a list of the slow and fast forms. With regard to mechanisms of genetic change, what features do slow and rapid speciation have in common? What features are different?
> The raw material for evolution is random mutation. Discuss whether or not you view evolution as a random process.
> What technique must be used to visualize a virus?
> Discuss how researchers determined that TMV is a virus that causes damage to plants.
> Chapter 21 describes a blotting method known as Northern blotting, which can be used to detect RNA transcribed from a particular gene or a particular operon. In this method, a specific RNA is detected by using a short segment of cloned DNA as a probe. Th
> Answer the following questions that pertain to the experiment of Figure 14.7. A. Why was β-ONPG used? Why was no yellow color observed in one of the four tubes? Can you propose alternative methods to measure the level of expression of the la
> What are the two enzymatic functions of reverse transcriptase?
> Researchers can introduce loss-of-function mutations into genes using the CRISPR/Cas9 technology described in Chapter 21. If you used this technology to produce the following homozygous loss-of-function mutations in a winter-annual strain of Arabidopsis,
> What is miRNA replacement therapy? Describe three examples of this treatment approach.
> Explain the rationale behind a DNase I footprinting experiment.
> In the technique of DNase I footprinting, the binding of a protein to a region of DNA protects that region from digestion by DNase I by blocking the ability of DNase I to gain access to the DNA. In the DNase I footprinting experiment shown here, a resear
> As described in Chapter 15 (Figures 15.7 and 15.8), certain regulatory transcription factors bind to DNA and activate RNA polymerase II. When glucocorticoid binds to the glucocorticoid receptor (a regulatory transcription factor), this changes the confor
> An electrophoretic mobility shift assay can be used to study the binding of proteins to a segment of DNA. In the experiment shown here, an EMSA was used to examine the requirements for the binding of RNA polymerase II (from eukaryotic cells) to the promo
> Certain hormones, such as epinephrine, can increase the levels of cAMP within cells. Let’s suppose you pretreat cells with or without epinephrine and then prepare a cell extract that contains the CREB protein (see Chapter 15 for a description of the CREB
> Describe the rationale behind the electrophoretic mobility shift assay.
> A cloned gene fragment contains a regulatory element that is recognized by a regulatory transcription factor. Previous experiments have shown that the presence of a hormone results in transcriptional activation by this transcription factor. To study this
> Explain the basis for using an antibody as a probe in a Western blotting experiment.
> What environmental conditions favor a switch to the lytic cycle?
> Let’s suppose a researcher was interested in the effects of mutations on the expression of a protein-encoding gene that encodes a polypeptide that is 472 amino acids in length. This polypeptide is expressed in leaf cells of Arabidopsis thaliana. Because
> If you wanted to know if a protein was made during a particular stage of development, what technique would you choose?
> Outline the general strategy used in metagenomics.
> What is meant by sequencing by synthesis?
> Discuss the advantages of next-generation sequencing technologies
> A bacterium has a genome size of 4.4 Mb. If a researcher carries out shotgun DNA sequencing and sequences a total of 19 Mb, what is the probability that a base will be left unsequenced? What percentage of the total genome will be left unsequenced?
> Describe how you would clone a gene by positional cloning. Explain how a (previously made) contig would make this task much easier.
> A human gene, which we will call gene X, is located on chromosome 11 and is found as a normal allele and a recessive disease causing allele. The location of gene X has been approximated on the map shown here, which contains four STSs, labeled STS-1, STS-
> Four cosmid clones, which we will call cosmids A, B, C, and D, were hybridized to each other in pairwise combinations. The insert size of each cosmid was also analyzed. The following results were obtained: Draw a map that shows the order of the inserts
> Take a look at question 3 in More Genetic TIPS and the codon table in Chapter 13. Assuming that a mutation causing a single base change is more likely than one causing a double base change, propose how the Asn-141, Ile-141, and Thr-141 codons arose. In y
> Let’s suppose a drug inhibits the function of the N protein. Would such a drug favor the lysogenic cycle, favor the lytic cycle, or prevent both cycles from occurring?
> What would you expect to be the minimum percentage of matching peaks in an automated DNA fingerprint for the following pairs of individuals? A. Mother and son B. Sister and brother C. Uncle and niece D. Grandfather and grandson
> As discussed in this chapter and Chapter 27, genes are sometimes transferred between different species via horizontal gene transfer. Discuss how horizontal gene transfer might lead to misleading results when constructing a phylogenetic tree. How could yo
> A homologous DNA region, which was 20,000 bp in length, was sequenced from four different species. The following numbers of nucleotide differences were obtained: Construct a phylogenetic tree that describes the evolutionary relationships among these fou
> Discuss how the principle of parsimony can be used in a cladistics approach to constructing a phylogenetic tree.
> A team of researchers has obtained a dinosaur bone (Tyrannosaurus rex) and has attempted to extract ancient DNA from it. Using primers for the 12S rRNA mitochondrial gene, they carried out PCR and obtained a DNA segment that had a sequence homologous to
> From the results of the experiment of Figure 29.13, explain how we know that the kiwis are more closely related to the emu and cassowary than to the moas. Cite particular regions in the sequences that support your answer. From Figure 29.13: Experime
> Prehistoric specimens often contain minute amounts of ancient DNA. What technique can be used to increase the amount of DNA in an older sample? Explain how this technique is performed and how it increases the amount of a specific region of DNA.
> Explain why molecular techniques were needed to provide evidence for the neutral theory of evolution.
> A species of antelope has 20 chromosomes per set. The species is divided by a mountain range into two separate populations, which we will call the eastern and western population. In a comparison of the karyotypes of these two populations, it was discover
> What is meant by the term emerging virus?
> F1 hybrids between two species of cotton, Gossypium barbadense and Gossypium hirsutum, are very vigorous plants. However, F1 crosses produce many seeds that do not germinate and a high percentage of very weak F2 offspring. Suggest two reasons for these o
> A researcher sequenced a portion of a bacterial gene and obtained the following sequence, beginning with the start codon, which is underlined: ATG CCG GAT TAC CCG GTC CCA AAC AAA ATG ATC GGC CGC CGA ATC TAT CCC The bacterial strain that contained this g
> Two diploid species of closely related frogs, which we will call species A and species B, were analyzed with regard to the genes that encode an enzyme called hexokinase. Species A has two distinct copies of this gene: A1 and A2. In other words, this dipl
> Sympatric speciation by allotetraploidy has been proposed as a common mechanism for speciation. Let’s suppose you were interested in the origin of certain grass species in southern California. Experimentally, how would you go about determining if some of
> Two populations of snakes are separated by a river. The snakes cross the river only on rare occasions. The snakes in the two populations look very similar to each other, except that the members of the population on the eastern bank of the river have a ye
> What is a biological control agent? Briefly describe two examples.
> Which of the following statements about molecular markers are true? A. All molecular markers are segments of DNA that carry specific genes. B. A molecular marker is a segment of DNA that is found at a specific location in a genome. C. We can follow th
> For each of the following, decide if it could be appropriately described as a genome: A. The E. coli chromosome B. Human chromosome 11 C. A complete set of 10 chromosomes in corn D. A copy of the single-stranded RNA packaged into human immunodeficien
> A person with a rare genetic disease has a sample of her chromosomes subjected to in situ hybridization using a probe that is known to recognize band p11 on chromosome 7. Even though her chromosomes look cytologically normal, the probe does not bind to t
> What is bioremediation? What is the difference between biotransformation and biodegradation?
> Which cycle produces new phage particles?
> A conjugation-deficient strain of A. radiobacter is used to combat crown gall disease. Explain how this bacterium prevents the disease, and describe the advantage of using a conjugation-deficient strain.
> What is a recombinant microorganism? Discuss examples.
> Make a list of the differences between the Holliday model and the double-strand break model.
> What is gene conversion?
> In the Holliday model for homologous recombination, the resolution steps can produce recombinant or nonrecombinant chromosomes. Explain how this can occur.
> What are recombinant chromosomes? How do they differ from the original parental chromosomes from which they are derived?
> Draw the structural feature of a dideoxyribonucleotide that causes chain termination. Explain how it does this.
> What is cDNA? In eukaryotes, how does cDNA differ from genomic DNA?
> Write a double-stranded DNA sequence that is 20 base pairs in length and is palindromic.
> What is a restriction enzyme? What structure does it recognize? What type of chemical bond does it cleave? Be as specific as possible.
> During which step of the reproductive cycle can a virus remain latent?
> According to the double-strand break model, does gene conversion necessarily involve DNA mismatch repair? Explain.
> What events does the RecA protein facilitate?
> In recombinant chromosomes, where is gene conversion likely to take place: near the breakpoint or far away from the breakpoint? Explain.
> Discuss three important advances that have resulted from gene cloning
> Is homologous recombination an example of mutation? Explain.
> What two molecular mechanisms can result in gene conversion? Do both occur in the double-strand break model?
> Which steps in the double-strand break model for recombination would be inhibited if the following proteins were missing? Explain the function of each protein required for the step that is inhibited. A. RecBCD B. RecA C. RecG D. RuvABC
> The molecular mechanism of SCE is similar to homologous recombination between homologs except that the two segments of DNA are sister chromatids instead of homologous chromatids. If branch migration occurs during SCE, will a heteroduplex be formed? Expla
> Describe the similarities and differences between homologous recombination involving sister chromatid exchange (SCE) and that involving homologs. Would you expect the same types of proteins to be involved in both processes? Explain.
> In Chapters 12 through 16, we discussed many sequences that are outside a coding sequence but are important for gene expression. Look up two of these sequences and write them out. Explain how a mutation could change these sequences, thereby altering gene
> If a mutation prevented IRP from binding to the IRE in the ferritin mRNA, how would the mutation affect the regulation of ferritin synthesis? Do you think there would be too much or too little ferritin?
> A point mutation occurs in the middle of the coding sequence for a gene. Which types of mutations—silent, missense, nonsense, and frameshift—would be most likely to disrupt protein function and which would be least likely?
> Is each of the following mutations a silent, missense, nonsense, or frameshift mutation? The original DNA strand is 5′–ATGGGACTAGATACC–3′. (Note: Only the coding strand is shown; the first codon is methionine.) A. 5′–ATGGGTCTAGATACC–3′ B. 5′–ATGCGACTAG
> Lactose permease is encoded by the lacY gene of the lac operon. Suppose a mutation occurred at codon 64 that changed the normal glycine codon into a valine codon. The mutant lactose permease is unable to function. However, a second mutation, which change
> X-rays strike a chromosome in a living cell and ultimately cause the cell to die. Did the X-rays produce a mutation? Explain why or why not.
> How would each of the following types of mutations affect protein function or the amount of functional protein that is expressed from a gene? A. Nonsense mutation B. Missense mutation C. Up promoter mutation D. Mutation that affects splicing
> What does a suppressor mutation suppress? What is the difference between an intragenic and an intergenic suppressor?
> Achondroplasia is a rare form of dwarfism. It is caused by an autosomal dominant mutation within a single gene. Among 1,422,000 live births, the number of babies born with achondroplasia was 31. Among those 31 babies, 18 of them had one parent with achon
> What is the difference between the mutation rate and the mutation frequency?
> With regard to TNRE, what is meant by the term anticipation?
> Trinucleotide repeat expansions (TNREs) are associated with several different human inherited diseases. Certain types of TNREs produce a long stretch of the amino acid glutamine within the encoded protein. When a TNRE exerts its detrimental effect by pro
> Why are insulators important for gene regulation in eukaryotes?
> What is the difference between an ancestral character and a shared derived character?
> If a mutagen causes bases to be removed from nucleotides within DNA, what repair system would fix this damage?

