Question: Transcription factors such as the glucocorticoid
Transcription Factors such as the glucocorticoid receptor and the CREB protein form homodimers and activate transcription. Other transcription factors form heterodimers. For example, a transcription factor known as myogenic bHLH forms a heterodimer with a protein called the E protein. This heterodimer activates the transcription of genes that promote muscle cell differentiation. However, when myogenic bHLH forms a heterodimer with a protein called the Id protein, transcriptional activation does not occur. (Note: Id stands for “Inhibitor of differentiation.â€) Which of the following possibilities best explains this observation? Only one possibility is correct.
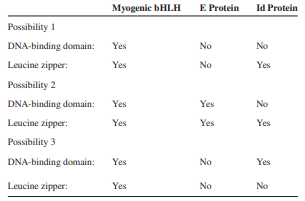
Transcribed Image Text:
Myogenic BHLH E Protein Id Protein Possibility 1 DNA-binding domain: Yes No No Leucine zipper: Yes No Yes Possibility 2 DNA-binding domain: Yes Yes No Leucine zipper: Yes Yes Yes Possibility 3 DNA-binding domain: Yes No Yes Leucine zipper: Yes No No
> What is an insulator? Describe two different ways that insulators may exert their effects.
> Histones are thought to be displaced as RNA polymerase is transcribing a gene. What would be the potentially harmful consequences if histones were not put back onto a gene after RNA polymerase had passed?
> What is a nucleosome-free region? Where are such regions typically found in a genome? How are nucleosome-free regions thought to be functionally important?
> What is meant by the term histone code? With regard to gene regulation, what is the proposed role of the histone code?
> An ncRNA may have the following functions: scaffold, guide, alterer of protein function or stability, ribozyme, blocker, and/or decoy. Which of those functions is/are mediated by each of the ncRNAs listed next? (Note: A single ncRNA may have more than on
> What are the two ways in which piRNAs and PIWI proteins prevent the movement of transposable elements?
> Explain how the acetylation of core histones may loosen chromatin packing.
> What is a histone variant?
> Explain how the VIN3/PRC2 complex specifically binds to the FLC gene.
> Explain how the miR-200 family of miRNAs behave as tumorsuppressor genes. What happens when their expression is blocked or decreased?
> List five types of cancer in which ncRNAs can be involved.
> Outline the steps that occur when piRISCs silence transposable elements by repressing transcription and by directly inhibiting TE RNAs. What is the role of piRNAs in this process?
> What are the roles of Cas1, Cas2, and Cas9 proteins in bacterial genome defense?
> In the CRISPR-Cas system, does the tracrRNA act as a scaffold, guide, ribozyme, blocker, decoy, and/or alterer of protein function or stability?
> Compare and contrast the roles of crRNA and tracrRNA in the defense process against bacteriophages provided by the CRISPRCas system.
> Look at Figure 17.6 and predict what would happen if the SRP RNA was unable to stimulate the GTPase activities of the GTPbinding proteins within SRP and the SRP receptor. From Figure 17.6: Ribosome 5' 3' ER signal sequence NH, As a polypeptide is b
> Which component of the CRISPR-Cas system directly recognizes the bacteriophage DNA?
> List and briefly describe four types of molecules that can bind to an ncRNA.
> In general, explain how epigenetic modifications are an important mechanism for developmental changes that lead to specialized body parts and cell types. How do the trithorax and polycomb group complexes participate in this process?
> Following X-chromosome inactivation, most of the genes on the inactivated X chromosome are silenced. Explain how. Name one gene that is not silenced.
> Outline the molecular steps in the process of X-chromosome inactivation (XCI). Which step plays a key role in choosing which of the X chromosomes will remain active and which will be inactivated?
> Let’s suppose a mutation removes the ICR next to the Igf2 gene. If this mutation is inherited from the mother, will the Igf2 gene (from the mother) be silenced or expressed? Explain.
> Explain how DNA methylation and the formation of a DNA loop control the expression of the Igf2 gene in mammals. How is this gene imprinted so that only the paternal copy is expressed in offspring?
> What is the key difference between cis- and trans-epigenetic mechanisms for maintaining an epigenetic modification? In Chapter 5, we considered genomic imprinting of the Igf2 gene, in which offspring express the copy of the gene they inherit from their f
> Explain how epigenetic changes may be targeted to specific genes.
> List and briefly describe five types of molecular mechanisms that may underlie epigenetic gene regulation.
> If a winter-annual strain of Arabidopsis is grown in a greenhouse and not exposed to cold temperatures, its ability to flower is inhibited. Which gene is responsible for this inhibition?
> Why is GTP necessary for this process?
> Is paramutation a cis- or a trans-epigenetic mechanism?
> How can environmental agents that do not cause gene mutations contribute to cancer? Would these epigenetic changes be passed to offspring?
> Using coat color in mice and the development of female honeybees as examples, explain how dietary factors can cause epigenetic modifications, leading to phenotypic effects.
> With regard to development, what would the dire consequences be if polycomb group complexes did not function properly?
> Describe the molecular steps by which polycomb group complexes cause epigenetic gene silencing.
> What are the contrasting roles of trithorax and polycomb group complexes during development in animals and plants?
> Define epigenetics. Are all epigenetic changes passed from parent to offspring? Explain.
> Let’s suppose a mutation in the glucocorticoid receptor does not prevent the binding of the glucocorticoid hormone to the protein but prevents the ability of the receptor to activate transcription. Make a list of all the possible defects that may explain
> Describe the steps that need to occur for the glucocorticoid receptor to bind to a GRE.
> The binding of a small effector molecule, protein-protein interactions, and covalent modifications are three common ways to modulate the activities of transcription factors. Which of these three mechanisms are used by steroid receptors and by the CREB pr
> Which type of snoRNA causes an rRNA to be methylated?
> What is a clade?
> Transcription factors usually contain one or more motifs that play key roles in their function. What is the function of the following motifs? A. Helix-turn-helix B. Zinc finger C. Leucine zipper
> Is each of the following statements true or false? A. An enhancer is a type of regulatory element. B. A core promoter is a type of regulatory element. C. Regulatory transcription factors bind to regulatory elements. D. An enhancer may cause the down
> What are the functions of transcriptional activator proteins and repressor proteins? Explain how they work at the molecular level.
> What is meant by the term transcription factor modulation? List three general ways this can occur.
> Discuss the structure and function of regulatory elements. Where are they located relative to the core promoter?
> Briefly describe three ways that ATP-dependent chromatin-remodeling complexes may change chromatin structure.
> The gene that encodes the enzyme called tyrosine hydroxylase is known to be activated by the CREB protein. Tyrosine hydroxylase is expressed in nerve cells and is involved in the synthesis of catecholamine, a neurotransmitter. The exposure of cells to ad
> The DNA-binding domain of each CREB protein subunit recognizes the sequence 5′–TGACGTCA–3′. Due to random chance, how often would you expect this sequence to occur in the human genome, which contains approximately 3 billion base pairs? Actually, only a f
> An enhancer, located upstream from a gene, has the following sequence: 5′–GTAG–3′ 3′–CATC–5′ This enhancer is orientation-independent. Which of the following sequences also works as an enhancer? A. 5′–CTAC–3′ 3′–GATG–5′ B. 5′–GATG–3′ 3′–CTAC–5′ C.
> Explain why RISC binds to a specific mRNA. What type of bonding occurs?
> The glucocorticoid receptor and the CREB protein are two examples of transcriptional activators. These proteins bind to response elements and activate transcription. (Note: The answers to this question are not directly described in this chapter. You have
> A particular drug inhibits the protein kinase that is responsible for phosphorylating the CREB protein. How would this drug affect the following events? A. The ability of the CREB protein to bind to CREs B. The ability of extracellular hormones to enha
> Explain how phosphorylation affects the function of the CREB protein.
> Discuss the common points of control in eukaryotic gene regulation.
> If an abnormal repressor protein could still bind allolactose but the binding of allolactose did not alter the conformation of the repressor protein, how would the expression of the lac operon be affected?
> In the lac operon, how would gene expression be affected if each one of the following segments was missing? A. lac operon promoter B. Operator site C. lacA gene
> What is enzyme adaptation? From a genetic point of view, how does it occur?
> Some mutations have a cis-effect, whereas others have a transeffect. Explain the molecular differences between cis- and transmutations. Which type of mutation (cis or trans) can be complemented in a merozygote experiment?
> An operon is repressible—a small effector molecule turns off its transcription. Which combination(s) of small effector molecule and regulatory protein could be involved in this process? A. An inducer plus a repressor B. A corepressor plus a repressor
> Transcriptional regulation often involves a regulatory protein that binds to a segment of DNA and a small effector molecule that binds to the regulatory protein. Do each of the following terms apply to a regulatory protein, a segment of DNA, or a small e
> What types of molecules can bind to a non-coding RNA?
> If a gene is repressible and under positive control, what kind of effector molecule and regulatory protein are involved in its regulation? Explain how the binding of the effector molecule affects the regulatory protein.
> Transcriptional repressor proteins (e.g., lac repressor), antisense RNA, and feedback inhibition are three different mechanisms that turn off the expression of genes and gene products. Which of these three mechanisms will be most effective in each of the
> How are the actions of lac repressor and trp repressor similar and how are they different with regard to their binding to operator sites, their effects on transcription, and the influences of small effector molecules?
> Using three examples, describe how allosteric sites are important in the function of genetic regulatory proteins.
> A species of bacteria can synthesize the amino acid histidine, so they do not require histidine in their growth medium. A key enzyme, which we will call histidine synthetase, is necessary for histidine biosynthesis. When these bacteria are given histidin
> In E. coli, a methyltransferase enzyme encoded by the dam gene recognizes the sequence 5′–GATC–3′ and attaches a methyl group to the nitrogen at position 6 of adenine. E. coli strains that have the dam gene deleted are known to have a higher spontaneous
> Discuss the similarities and differences between nucleotide excision repair and the mismatch repair system.
> Three common ways to repair changes in DNA structure are nucleotide excision repair, mismatch repair, and homologous recombination repair. Which of these three mechanisms would be used to fix the following types of DNA changes? A. A change in the struct
> What is the underlying genetic defect that causes xeroderma pigmentosum? How can the symptoms of this disease be explained by the genetic defect?
> When DNA N-glycosylase recognizes a thymine dimer, it detects only the thymine located on the 5′ side of the dimer as being abnormal. Make a drawing and explain the steps whereby a thymine dimer is repaired by the consecutive actions of DNA N-glycosylase
> Are queen and worker bees genetically different from each other?
> With regard to the repair of double-strand breaks, what are the advantages and disadvantages of homologous recombination repair versus nonhomologous end joining?
> What are the two main mechanisms by which cells repair doublestrand breaks? Briefly describe each one.
> During mismatch repair, why is it necessary to distinguish between the template strand and the newly made daughter strand? How is this accomplished?
> How would nucleotide excision repair be affected if one of the following proteins was missing? Describe the condition of the DNA if the repair was attempted in the absence of the protein. A. UvrA B. UvrC C. UvrD D. DNA polymerase
> Explain how alternative splicing affects sex determination in Drosophila.
> Predict the phenotypic consequences of each of the following mutations: A. apetala1 defective B. pistillata defective C. apetala1 and pistillata defective
> Let’s suppose the mutation rate for converting a B allele into a b allele is 10–4. The current allele frequencies are B = 0.6 and b = 0.4. How long will it take for the allele frequencies to equal each other, assuming that no genetic drift takes place?
> Antibiotics are commonly used to combat bacterial and fungal infections. During the past several decades, however, antibioticresistant strains of microorganisms have become alarmingly prevalent. This has undermined the effectiveness of antibiotics in tre
> A family pedigree is shown here. A. What is the inbreeding coefficient for individual IV-2? Who is/ are her parents’ common ancestor(s)? B. Based on the data shown in this pedigree, is individual III-4 inbred? 1-1 I-2 Il-1 Il-2 II
> A family pedigree is shown here. A. What is the inbreeding coefficient for individual IV-3? B. Based on the data shown in this pedigree, is individual IV-4 inbred? 1-2 1-3 II-1 II-2 II-3 Il-4 Il-5 Il-6 III-1 III-2 III-3 III-4 III-5 III-6 III-7 III-
> In the F1 offspring, what happened to the B-I allele that was inherited from the parent at the top right?
> Using the pedigree shown here, answer the following questions for individual VI-1. A. Is this individual inbred? B. If so, who is/are her parents’ common ancestor(s)? C. Calculate the inbreeding coefficient for VI-1. D. Are the pare
> With regard to heterosis, is each of the following statements consistent with the dominance hypothesis, the overdominance hypothesis, or both? A. Strains that have been highly inbred have become monomorphic for one or more recessive alleles that are som
> If you were comparing the karyotypes of species that are closely related evolutionarily, what types of similarities and differences would you expect to find?
> As discussed in Chapter 27, genetic variation is prevalent in natural populations. This variation is revealed in the DNA sequencing of genes. Based on the neutral theory of evolution, discuss the relative importance of natural selection against detriment
> For each of the following examples, discuss whether the observed result is due to neutral mutations or mutations that have been acted on by natural selection, or both: A. When comparing sequences of homologous genes, differences in the coding sequence a
> Compare and contrast the neutral theory of evolution and the Darwinian (i.e., selectionist) theory of evolution. Explain why the neutral theory of evolution is sometimes called non-Darwinian evolution.
> Take a look at the α-globin and β-globin amino acid sequences in Figure 29.11. Which sequences are more similar, the α globin in humans and the α globin in horses, or the α globin in hum
> Plant seeds contain storage proteins that are encoded by the plant’s genes. When a seed germinates, these proteins are rapidly hydrolyzed (i.e., the covalent bonds between amino acids within the polypeptides are broken), which releases amino acids for th
> When comparing the coding regions of a protein-encoding gene among closely related species, certain regions are commonly found to have evolved more rapidly (i.e., have tolerated more changes in sequence) than other regions. Explain why different regions
> Which would you expect to exhibit a faster rate of evolutionary change, the nucleotide sequence of a gene or the amino acid sequence of the encoded polypeptide of the same gene? Explain your answer.
> Describe how the compaction of nucleosomes into a knot-like structure could silence gene expression.
> Would the rate of deleterious or beneficial mutations be a good molecular clock? Why or why not?
> What is meant by the term molecular clock? How is this concept related to the neutral theory of evolution?
> The following are DNA sequences from two homologous genes: TTGCATAGGCATACCGTATGATATCGAAAACTAGAAAAATAGGGCGATAGCTA GTATGTTATCGAAAAGTAGCAAAATAGGGCGATAGCTACCCAGACTACCGGAT The two sequences, however, do not begin and end at the same location. Try to line the
> Discuss the major differences among allopatric, parapatric, and sympatric speciation.
> Discuss whether the phenomenon of reproductive isolation applies to bacteria, which reproduce asexually. How would a geneticist divide bacteria into separate species?
> Alloploids are produced by crosses involving two different species. Explain why alloploids may be reproductively isolated from the two original species from which they were derived. Explain why alloploids are usually sterile, whereas allotetraploids (con

